|
The frog genus Xenopus is widely used as a model system for studying developmental biology and fundamental cell biological processes. The advantages that Xenopus offer as an experimental system include (1) the availability of large abundant eggs that are easily manipulated, (2) ready accessibility to any developmental stage, and (3) conservation of cellular pathways between Xenopus and mammals. Over the past 50 years, pioneering studies on Xenopus have been crucial towards our understanding of nuclear reprogramming, embryonic patterning, membrane channels and receptors, and the cell cycle. Despite its popularity for biomedical research, genomic resources for Xenopus have been lagging behind other model organisms. A major reason for this is that early developmental and molecular studies have relied on a particular species known as Xenopus laevis, which is pseudotetraploid as a result of genome duplication around 30 million years ago. The presence of four copies of every gene complicates genetic and genomic research. Fortunately, a closely related frog, Xenopus tropicalis, has a more amenable diploid genome and it also retains the same advantages as Xenopus laevis. Although the genome of Xenopus tropicalis has been sequenced and published, other genomic tools and resources are still needed. We present on this website the developmental transcriptome of Xenopus tropicalis from a two-cell fertilized embryo to a feeding tadpole. |
This webpage presents the data accompanying our paper: Meng How Tan, Kin Fai Au, Arielle L. Yablonovitch, Andrea E. Wills, Jason Chuang, Julie C. Baker, Wing Hung Wong, and Jin Billy Li Interactive visualizations on this webpage require one of the following web browsers: |
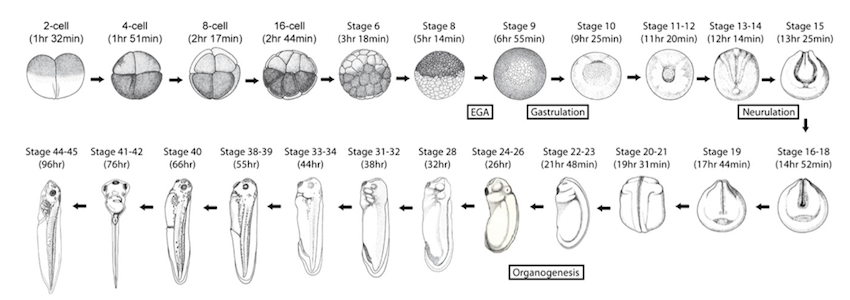
The developmental stages investigated in this study. Image is adapted from Nieuwkoop and Faber 1994 and reprinted with permission from
Garland Science/Taylor & Francis LLC © 1994.



